| Index to this page |
Enzymes are catalysts. Most are proteins. (A few ribonucleoprotein enzymes have been discovered and, for some of these, the catalytic activity is in the RNA part rather than the protein part. Link to discussion of these ribozymes.)
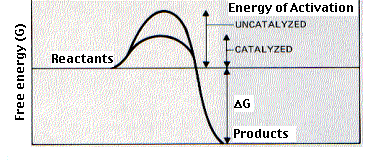
Enzymes bind temporarily to one or more of the reactants of the reaction they catalyze. In doing so, they lower the amount of activation energy needed and thus speed up the reaction. |
2H2O2 -> 2H2O + O2
One molecule of catalase can break 40 million molecules of hydrogen peroxide each second.
CO2 + H2O <=> H2CO3
It enables red blood cells to transport carbon dioxide from the tissues to the lungs. [Discussion]
One molecule of carbonic anhydrase can process one million molecules of CO2 each second.
Acetylcholinesterase. It catalyzes the breakdown of the neurotransmitter acetylcholine at several types of synapses as well as at the neuromuscular junction — the specialized synapse that triggers the contraction of skeletal muscle.
One molecule of acetylcholinesterase breaks down 25,000 molecules of acetylcholine each second. This speed makes possible the rapid "resetting" of the synapse for transmission of another nerve impulse.
| Enzyme activity can be analyzed quantitatively. Some of the ways this is done are described in the page Enzyme Kinetics. Link to it. |
In order to do its work, an enzyme must unite — even if ever so briefly — with at least one of the reactants. In most cases, the forces that hold the enzyme and its substrate are noncovalent, an assortment of:
| Link to discussion of the noncovalent forces that hold macromolecules together. |
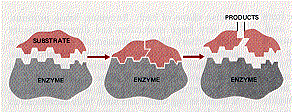
Most of these interactions are weak and especially so if the atoms involved are farther than about one angstrom from each other. So successful binding of enzyme and substrate requires that the two molecules be able to approach each other closely over a fairly broad surface. Thus the analogy that a substrate molecule binds its enzyme like a key in a lock.
This requirement for complementarity in the configuration of substrate and enzyme explains the remarkable specificity of most enzymes. Generally, a given enzyme is able to catalyze only a single chemical reaction or, at most, a few reactions involving substrates sharing the same general structure.
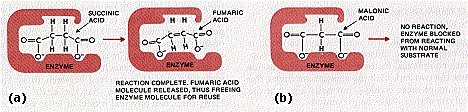
The necessity for a close, if brief, fit between enzyme and substrate explains the phenomenon of competitive inhibition.
One of the enzymes needed for the release of energy within the cell is succinic dehydrogenase.
It catalyzes the oxidation (by the removal of two hydrogen atoms) of succinic acid (a). If one adds malonic acid to cells, or to a test tube mixture of succinic acid and the enzyme, the action of the enzyme is strongly inhibited. This is because the structure of malonic acid allows it to bind to the same site on the enzyme (b). But there is no oxidation so no speedy release of products. The inhibition is called competitive because if you increase the ratio of succinic to malonic acid in the mixture, you will gradually restore the rate of catalysis. At a 50:1 ratio, the two molecules compete on roughly equal terms for the binding (=catalytic) site on the enzyme.
Coenzymes may be covalently bound to the protein part (called the apoenzyme) of enzymes as a prosthetic group. Others bind more loosely and, in fact, may bind only transiently to the enzyme as it performs its catalytic act.
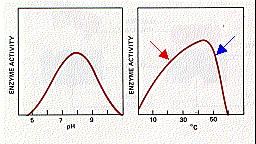 The activity of enzymes is strongly affected by changes in
pH and temperature. Each enzyme works best at a certain
pH (left graph) and temperature (right graph), its activity decreasing
at values above and below that point. This is not surprising
considering the importance of
The activity of enzymes is strongly affected by changes in
pH and temperature. Each enzyme works best at a certain
pH (left graph) and temperature (right graph), its activity decreasing
at values above and below that point. This is not surprising
considering the importance of
Changes in pH alter the state of ionization of charged amino acids (e.g., Asp, Lys) that may play a crucial role in substrate binding and/or the catalytic action itself. Without the unionized -COOH group of Glu-35 and the ionized -COO- of Asp-52, the catalytic action of lysozyme would cease.
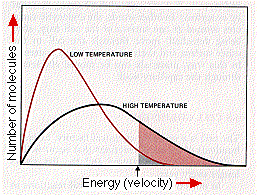
Hydrogen bonds are easily disrupted by increasing temperature. This, in turn, may disrupt the shape of the enzyme so that its affinity for its substrate diminishes. The ascending portion of the temperature curve (red arrow in right-hand graph above) reflects the general effect of increasing temperature on the rate of chemical reactions (graph at left). The descending portion of the curve above (blue arrow) reflects the loss of catalytic activity as the enzyme molecules become denatured at high temperatures.
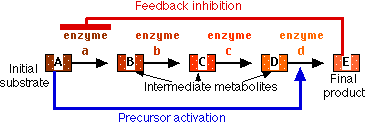
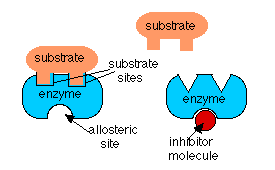
In the case if feedback inhibition and precursor activation, the activity of the enzyme is being regulated by a molecule which is not its substrate. In these cases, the regulator molecule binds to the enzyme at a different site than the one to which the substrate binds. When the regulator binds to its site, it alters the shape of the enzyme so that its activity is changed. This is called an allosteric effect.
The four mechanisms described above regulate the activity of enzymes already present within the cell.
What about enzymes that are not needed or are needed but not present?
Here, too, control mechanisms are at work that regulate the rate at which new enzymes are synthesized. Most of these controls work by turning on — or off — the transcription of genes.
If, for example, ample quantities of an amino acid are already available to the cell from its extracellular fluid, synthesis of the enzymes that would enable the cell to produce that amino acid for itself is shut down.
Conversely, if a new substrate is made available to the cell, it may induce the synthesis of the enzymes needed to cope with it. Yeast cells, for example, do not ordinarily metabolize lactose and no lactase can be detected in them. However, if grown in a medium containing lactose, they soon begin synthesizing lactase — by transcribing and translating the necessary gene(s) — and so can begin to metabolize the sugar.