| Index to this page |
The problem: How does a particular sequence of nucleotides specify a particular sequence of amino acids?
The answer: by means of transfer RNA molecules, each specific for one amino acid and for a particular triplet of nucleotides in mRNA called a codon. The family of tRNA molecules enables the codons in a mRNA molecule to be translated into the sequence of amino acids in the protein.
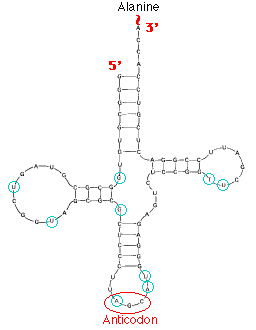
This image shows the structure of alanine transfer RNA (tRNAala) from yeast. It consists of a single strand of 77 ribonucleotides. The chain is folded on itself, and many of the bases pair with each other forming four helical regions. Loops are formed in the unpaired regions of the chain. (The bases circled in blue have been chemically-modified following synthesis of the molecule.)
At least one kind of tRNA is present for each of the 20 amino acids used in protein synthesis. (Some amino acids employ the services of two or three different tRNAs, so most cells contain as many as 32 different kinds of tRNA.) The amino acid is attached to the appropriate tRNA by an activating enzyme (one of 20 aminoacyl-tRNA synthetases) specific for that amino acid as well as for the tRNA assigned to it.
Each kind of tRNA has a sequence of 3 unpaired nucleotides — the anticodon — which can bind, following the rules of base pairing, to the complementary triplet of nucleotides — the codon — in a messenger RNA (mRNA) molecule. Just as DNA replication and transcription involve base pairing of nucleotides running in opposite direction, so the reading of codons in mRNA (5' -> 3') requires that the anticodons bind in the opposite direction. Anticodon: 3' CGA 5'
Codon: 5' GCU 3'
| U | C | A | G | ||
|---|---|---|---|---|---|
| U | UUU Phenylalanine (Phe) | UCU Serine (Ser) | UAU Tyrosine (Tyr) | UGU Cysteine (Cys) | U |
| UUC Phe | UCC Ser | UAC Tyr | UGC Cys | C | |
| UUA Leucine (Leu) | UCA Ser | UAA STOP | UGA STOP | A | |
| UUG Leu | UCG Ser | UAG STOP | UGG Tryptophan (Trp) | G | |
| C | CUU Leucine (Leu) | CCU Proline (Pro) | CAU Histidine (His) | CGU Arginine (Arg) | U |
| CUC Leu | CCC Pro | CAC His | CGC Arg | C | |
| CUA Leu | CCA Pro | CAA Glutamine (Gln) | CGA Arg | A | |
| CUG Leu | CCG Pro | CAG Gln | CGG Arg | G | |
| A | AUU Isoleucine (Ile) | ACU Threonine (Thr) | AAU Asparagine (Asn) | AGU Serine (Ser) | U |
| AUC Ile | ACC Thr | AAC Asn | AGC Ser | C | |
| AUA Ile | ACA Thr | AAA Lysine (Lys) | AGA Arginine (Arg) | A | |
| AUG Methionine (Met) or START | ACG Thr | AAG Lys | AGG Arg | G | |
| G | GUU Valine Val | GCU Alanine (Ala) | GAU Aspartic acid (Asp) | GGU Glycine (Gly) | U |
| GUC (Val) | GCC Ala | GAC Asp | GGC Gly | C | |
| GUA Val | GCA Ala | GAA Glutamic acid (Glu) | GGA Gly | A | |
| GUG Val | GCG Ala | GAG Glu | GGG Gly | G |
Note:
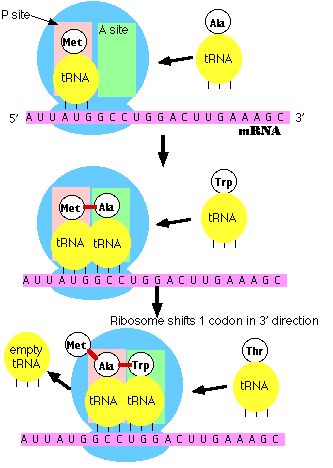
Note: the initiator tRNA is the only member of the tRNA family that can bind directly to the P site. The P site is so-named because, with the exception of initiator tRNA, it binds only to a peptidyl-tRNA molecule; that is, a tRNA with the growing peptide attached.
The A site is so-named because it binds only to the incoming aminoacyl-tRNA; that is the tRNA bringing the next amino acid. So, for example, the tRNA that brings Met into the interior of the polypeptide can bind only to the A site.
| (With a few special exceptions: link to mitochondrial genes and to nonstandard amino acids.) |
| External Link |
| Link to John Kyrk's superb animation of the entire process. |
| Please let me know by e-mail if you find a broken link in my pages.) |
Nonstop transcripts occur when there is no STOP codon in the message. As a result the ribosome is unable to recruit the release factors needed to leave the mRNA.
Nonstop transcripts are formed during RNA processing, e.g., by having the poly(A) tail put on before the STOP codon is reached.Eukaryotes and prokaryotes handle the problem of no STOP codon differently.
The expression of most genes is controlled at the level of their transcription. Transcription factors (proteins) bind to promoters and enhancers turning on (or off) the genes they control.
| Link to an example. |
However, there are some cases where gene expression is controlled at the level of translation instead.
| Link to a discussion. |
In many bacteria (and some eukaryotes), it turns out that the regulation of the level of certain metabolites is controlled by riboswitches. A riboswitch is a part of a molecule of messenger RNA (mRNA) with a specific binding site for the metabolite (or a close relative).
Examples:Other riboswitches act on transcription rather than translation. [Link]
It has been suggested that these regulatory mechanisms, which do not involve any protein, are a relict from an "RNA world".
Translation of at least one mRNA in humans is repressed by a protein — aminoacyl tRNA synthetase. In response to the inflammatory cytokine interferon-gamma [IFN-γ], the synthetase abandons its normal function (adding Glu and Pro to their respective tRNAs) and instead binds to the mRNA blocking its translation.
In some bacteria, a protein product may inhibit the further translation of its own mRNA (a kind of feedback inhibition). It does so by binding to a site which blocks the mRNA from further association with a ribosome.
In eukaryotes, the processes of transcription and translation are separated both spatially and in time. Transcription of DNA into mRNA occurs in the nucleus. Translation of mRNA into polypeptides occurs on polysomes in the cytoplasm.
In prokaryotes (which have no nucleus), both these steps of gene expression occur simultaneously: the nascent mRNA molecule begins to be translated even before its transcription from DNA is complete.
| View an electron micrograph showing polysomes formed during simultaneous transcription and translation in E. coli. |
| Recent evidence (reported by Iborra, et. al, in the 10 August 2001 issue of Science) shows that the distinction between prokaryotes and eukaryotes is not absolute. They find that 10 to 15% of translation in mammalian cells occurs in the nucleus, and that at least some of this translation occurs as the mRNA is still being synthesized by RNA polymerase (just as in E. coli) |
| Welcome&Next Search |